Multichannel Timeseries#
Show code cell source
from IPython.display import HTML
HTML("""
<div style="display: flex; justify-content: center; padding: 10px;">
<iframe width="560" height="315" src="https://www.youtube.com/embed/8oeuPptWPt8?si=yh-4iJYDfYpJZ7MQ" title="YouTube video player" frameborder="0" allow="accelerometer; clipboard-write; encrypted-media; gyroscope; picture-in-picture; web-share" referrerpolicy="strict-origin-when-cross-origin" allowfullscreen></iframe>
</div>
""")
Prerequisites#
What? |
Why? |
|---|---|
For context and workflow selection/feature guidance |
Overview#
For an introduction, please visit the ‘Index’ page. This workflow is tailored for processing and analyzing ‘medium-sized’ multichannel timeseries data derived from electrophysiological recordings.
What Defines a ‘Medium-Sized’ Dataset?#
In this context, we’ll define a medium-sized dataset as that which is challenging for browsers (roughly more than 100,000 samples) but can be handled within the available RAM without exhausting system resources.
Why Downsample?#
Medium-sized datasets can strain the processing capabilities when visualizing or analyzing data directly in the browser. To address this challenge, we will employ a smart-downsampling approach - reducing the dataset size by selectively subsampling the data points. Specifically, we’ll make use of a variant of a downsampling algorithm called Largest Triangle Three Buckets (LTTB). LTTB allows data points not contributing significantly to the visible shape to be dropped, reducing the amount of data to send to the browser but preserving the appearance (and particularly the envelope, i.e. highest and lowest values in a region). This ensures efficient data handling and visualization without significant loss of information.
Downsampling is particularly beneficial when dealing with numerous timeseries sharing a common time index, as it allows for a consolidated slicing operation across all series, significantly reducing the computational load and enhancing responsiveness for interactive visualization. We’ll make use of a Pandas index to represent the time index across all timeseries.
Quick Introduction to MNE#
MNE (MNE-Python) is a powerful open-source Python library designed for handling and analyzing data like EEG and MEG. In this workflow, we’ll utilize an EEG dataset, so we demonstrate how to use MNE for loading, preprocessing, and conversion to a Pandas DataFrame. However, the data visualization section is highly generalizable to dataset types beyond the scope of MNE, so you can meet us there if you have your timeseries data as a Pandas DataFrame with a time index and channel columns.
Imports and Configuration#
from pathlib import Path
import holoviews as hv
import mne
import pandas as pd
import panel as pn
import pooch
from holoviews.operation.downsample import downsample1d
pn.extension('tabulator')
hv.extension('bokeh')
Loading and Inspecting the Data#
Let’s get some data! This section walks through obtaining an EEG dataset (2.6 MB).
DATA_URL = 'https://datasets.holoviz.org/eeg/v1/S001R04.edf'
DATA_DIR = Path('./data')
DATA_FILENAME = Path(DATA_URL).name
DATA_PATH = DATA_DIR / DATA_FILENAME
Note
If you are viewing this notebook as a result of using the `anaconda-project run` command, the data has already been ingested, as configured in the associated yaml file. Running the following cell should find that data and skip any further download.DATA_DIR.mkdir(parents=True, exist_ok=True)
# Download the data if it doesn't exist
if not DATA_PATH.exists():
print(f'Downloading data to: {DATA_PATH}')
pooch.retrieve(
url=DATA_URL,
known_hash=None,
fname=DATA_FILENAME,
path=DATA_DIR,
progressbar=True,
)
else:
print(f'Data exists at: {DATA_PATH}')
Data exists at: data/S001R04.edf
Once we have a local copy of the data, the next crucial step is to load it into an analysis-friendly format and inspect its basic characteristics:
raw = mne.io.read_raw_edf(DATA_PATH, preload=True)
print('num samples in dataset:', len(raw.times) * len(raw.ch_names))
raw.info
Extracting EDF parameters from /home/runner/work/examples/examples/multichannel_timeseries/data/S001R04.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 19999 = 0.000 ... 124.994 secs...
num samples in dataset: 1280000
| General | ||
|---|---|---|
| MNE object type | Info | |
| Measurement date | 2009-08-12 at 16:15:00 UTC | |
| Participant | X | |
| Experimenter | Unknown | |
| Acquisition | ||
| Sampling frequency | 160.00 Hz | |
| Channels | ||
| EEG | ||
| Head & sensor digitization | Not available | |
| Filters | ||
| Highpass | 0.00 Hz | |
| Lowpass | 80.00 Hz | |
This step confirms the successful loading of the data and provides an initial understanding of its structure, such as the number of channels and samples.
Now, let’s preview the channel names, types, unit, and signal ranges. This describe method is from MNE, and we can have it return a Pandas DataFrame, from which we can sample some rows.
raw.describe(data_frame=True).sample(5)
| name | type | unit | min | Q1 | median | Q3 | max | |
|---|---|---|---|---|---|---|---|---|
| ch | ||||||||
| 58 | Po4. | eeg | V | -0.000184 | -0.000033 | -0.000001 | 0.000032 | 0.000204 |
| 25 | Af3. | eeg | V | -0.000348 | -0.000051 | -0.000008 | 0.000032 | 0.000511 |
| 20 | Cp6. | eeg | V | -0.000199 | -0.000028 | -0.000001 | 0.000026 | 0.000160 |
| 54 | P8.. | eeg | V | -0.000149 | -0.000027 | -0.000002 | 0.000023 | 0.000158 |
| 47 | P5.. | eeg | V | -0.000167 | -0.000033 | 0.000000 | 0.000033 | 0.000219 |
Pre-processing the Data#
Noise Reduction via Averaging#
Significant noise reduction is often achieved by employing an average reference, which involves calculating the mean signal across all channels at each time point and subtracting it from the individual channel signals:
raw.set_eeg_reference("average")
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
| General | ||
|---|---|---|
| Filename(s) | S001R04.edf | |
| MNE object type | RawEDF | |
| Measurement date | 2009-08-12 at 16:15:00 UTC | |
| Participant | X | |
| Experimenter | Unknown | |
| Acquisition | ||
| Duration | 00:02:05 (HH:MM:SS) | |
| Sampling frequency | 160.00 Hz | |
| Time points | 20,000 | |
| Channels | ||
| EEG | ||
| Head & sensor digitization | Not available | |
| Filters | ||
| Highpass | 0.00 Hz | |
| Lowpass | 80.00 Hz | |
Standardizing Channel Names#
From the output of the describe method, it looks like the channels are from commonly used standardized locations (e.g. ‘Cz’), but contain some unnecessary periods, so let’s clean those up to ensure smoother processing and analysis.
raw.rename_channels(lambda s: s.strip("."));
Optional: Enhancing Channel Metadata#
Visualizing physical locations of EEG channels enhances interpretative analysis. MNE has functionality to assign locations of the channels based on their standardized channel names, so we can go ahead and assign a commonly used arrangement (or ‘montage’) of electrodes (‘10-05’) to this data. Read more about making and setting the montage here.
montage = mne.channels.make_standard_montage("standard_1005")
raw.set_montage(montage, match_case=False)
| General | ||
|---|---|---|
| Filename(s) | S001R04.edf | |
| MNE object type | RawEDF | |
| Measurement date | 2009-08-12 at 16:15:00 UTC | |
| Participant | X | |
| Experimenter | Unknown | |
| Acquisition | ||
| Duration | 00:02:05 (HH:MM:SS) | |
| Sampling frequency | 160.00 Hz | |
| Time points | 20,000 | |
| Channels | ||
| EEG | ||
| Head & sensor digitization | 67 points | |
| Filters | ||
| Highpass | 0.00 Hz | |
| Lowpass | 80.00 Hz | |
We can see that the ‘digitized points’ (locations) are now added to the raw data.
Now let’s plot the channels using MNE plot_sensors on a top-down view of a head. Note, we’ll tweak the reference point so that all the points are contained within the depiction of the head.
sphere=(0, 0.015, 0, 0.099) # manually adjust the y origin coordinate and radius
raw.plot_sensors(show_names=True, sphere=sphere, show=False);
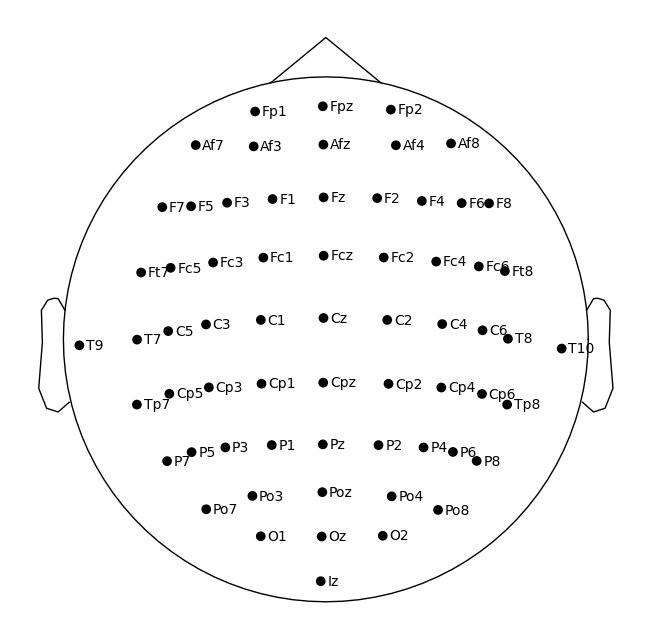
Data Visualization#
Preparing Data for Visualization#
We’ll use an MNE method, to_data_frame, to create a Pandas DataFrame. By default, MNE will convert EEG data from Volts to microVolts (µV) during this operation.
df = raw.to_data_frame()
df.set_index('time', inplace=True)
df.head()
| Fc5 | Fc3 | Fc1 | Fcz | Fc2 | Fc4 | Fc6 | C5 | C3 | C1 | ... | P8 | Po7 | Po3 | Poz | Po4 | Po8 | O1 | Oz | O2 | Iz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| time | |||||||||||||||||||||
| 0.00000 | 8.703125 | 15.703125 | 50.703125 | 52.703125 | 43.703125 | 39.703125 | -2.296875 | -0.296875 | 17.703125 | 31.703125 | ... | -7.296875 | 5.703125 | -21.296875 | -31.296875 | -52.296875 | -25.296875 | -19.296875 | -34.296875 | -25.296875 | -25.296875 |
| 0.00625 | 46.062500 | 34.062500 | 59.062500 | 56.062500 | 43.062500 | 36.062500 | 3.062500 | 22.062500 | 31.062500 | 33.062500 | ... | 8.062500 | 18.062500 | -9.937500 | -6.937500 | -25.937500 | 6.062500 | 37.062500 | 16.062500 | 27.062500 | 24.062500 |
| 0.01250 | -13.656250 | -14.656250 | 4.343750 | -1.656250 | 0.343750 | 8.343750 | -3.656250 | -24.656250 | -7.656250 | -1.656250 | ... | 46.343750 | 41.343750 | 13.343750 | 28.343750 | 15.343750 | 45.343750 | 43.343750 | 21.343750 | 34.343750 | 36.343750 |
| 0.01875 | -3.015625 | -4.015625 | 12.984375 | -2.015625 | 2.984375 | 7.984375 | -5.015625 | 0.984375 | 9.984375 | 8.984375 | ... | 23.984375 | 2.984375 | -15.015625 | -1.015625 | -5.015625 | 21.984375 | 18.984375 | 0.984375 | 28.984375 | 19.984375 |
| 0.02500 | 20.656250 | 10.656250 | 32.656250 | 12.656250 | 11.656250 | 2.656250 | -17.343750 | 13.656250 | 15.656250 | 12.656250 | ... | 21.656250 | 10.656250 | -4.343750 | 11.656250 | 2.656250 | 28.656250 | 5.656250 | -4.343750 | 31.656250 | 20.656250 |
5 rows × 64 columns
Creating the Multichannel Timeseries Plot#
As of the time of writing, there’s no easy way to track units with Pandas, so we can use a modular HoloViews approach to create and annotate dimensions with a unit, and then refer to these dimensions when plotting. Read more about annotating data with HoloViews here.
time_dim = hv.Dimension("time", unit="s") # match the df index name, 'time'
Now we will loop over the columns (channels) in the dataframe, creating a HoloViews Curve element from each. Since each column in the df has a different channel name, which is generally not describing a measurable quantity, we will map from the channel to a common amplitude dimension (see this issue for details of this enhancement for ‘wide’ tabular data), and collect each Curve element into a Python dictionary.
In configuring these curves, we apply the .opts method from HoloViews to fine-tune the visualization properties of each curve. The subcoordinate_y setting is pivotal for managing time-aligned, amplitude-diverse plots. When enabled, it arranges each curve along its own segment of the y-axis within a single composite plot. This method not only aids in differentiating the data visually but also in analyzing comparative trends across multiple channels, ensuring that each channel’s data is individually accessible and comparably presentable, thereby enhancing the analytical value of the visualizations. Applying subcoordinate_y has additional effects, such as creating a Y-axis zoom tool that applies to individual subcoordinate axes rather than the global Y-axis. Read more about subcoordinate_y here.
curves = {}
for col in df.columns:
col_amplitude_dim = hv.Dimension(col, label='amplitude', unit="µV") # map amplitude-labeled dim per chan
curves[col] = hv.Curve(df, time_dim, col_amplitude_dim, group='EEG', label=col)
# Apply options
curves[col] = curves[col].opts(
subcoordinate_y=True, # Essential to create vertically stacked plot
subcoordinate_scale=3,
color="black",
bgcolor="whitesmoke",
line_width=1,
hover_tooltips = [
("type", "$group"),
("channel", "$label"),
("time"),
("amplitude")],
tools=['xwheel_zoom'],
active_tools=["box_zoom"]
)
Using a HoloViews container, we can now overlay all the curves on the same plot.
curves_overlay = hv.Overlay(curves, kdims=[time_dim, 'Channel'])
overlay_opts = dict(ylabel="Channel",
show_legend=False,
padding=0,
min_height=600,
responsive=True,
title="",
)
curves_overlay = curves_overlay.opts(**overlay_opts)
Apply Downsampling#
Since there are dozens of channels and over a million data samples, we’ll make use of downsampling before trying to send all that data to the browser. We can use downsample1d imported from HoloViews. Starting in HoloViews version 1.19.0, integration with the tsdownsample library introduces enhanced downsampling algorithms. Read more about downsampling here.
curves_overlay_lttb = downsample1d(curves_overlay, algorithm='minmax-lttb')
Display Multichannel Timeseries Plot#
It is recommended to only have a single rendered instance of the same application component in a notebook, so we’ve commented out the following cell so we can demonstrate additional extensions below. But feel free to uncomment and display the following if you don’t want any further extensions.
# curves_overlay_lttb
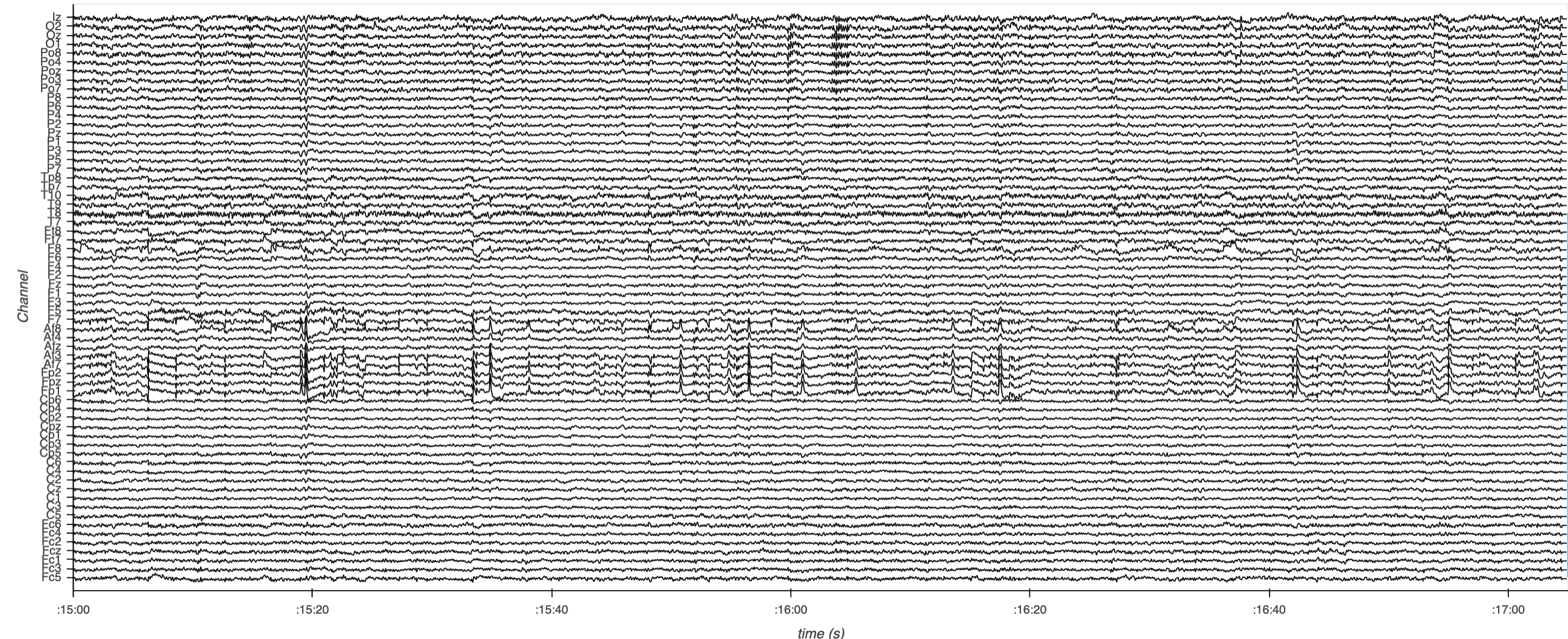
Here’s a static snapshot of what the previous cell produces (a DynamicMap of downsampled and overlaid curve subplots). 👉
print(curves_overlay_lttb)
:DynamicMap []
Minimap Extension#
To assist in navigating the dataset, we integrate a minimap widget. This secondary minimap plot provides a condensed overview of the entire dataset, allowing users to select and zoom into areas of interest quickly in the main plot while maintaining the contextualization of the zoomed out view.
We will employ datashader rasterization of the image for the minimap plot to display a browser-friendly, aggregated view of the entire dataset. Read more about datashder rasterization via HoloViews here.
from scipy.stats import zscore
from holoviews.operation.datashader import rasterize
from holoviews.plotting.links import RangeToolLink
channels = df.columns
time = df.index.values
y_positions = range(len(channels))
yticks = [(i, ich) for i, ich in enumerate(channels)]
z_data = zscore(df, axis=0).T
minimap = rasterize(hv.Image((time, y_positions, z_data), [time_dim, "Channel"], "amplitude"))
minimap_opts = dict(
cmap="RdBu_r",
colorbar=False,
xlabel='',
alpha=0.5,
yticks=[yticks[0], yticks[-1]],
toolbar='disable',
height=120,
responsive=True,
cnorm='eq_hist',
)
minimap = minimap.opts(**minimap_opts)
The connection between the main plot and the minimap is facilitated by a RangeToolLink, enhancing user interaction by synchronizing the visible range of the main plot with selections made on the minimap. Optionally, we’ll also constrain the initially displayed x-range view to a third of the duration.
RangeToolLink(minimap, curves_overlay_lttb, axes=["x", "y"],
boundsx=(0, time[len(time)//3]), # limit the initial selected x-range of the minimap
boundsy=(-.5,len(channels)//3), # limit the initial selected y-range of the minimap
use_handles=False, # Optionally, disable handles on the RangeTool box selection
)
RangeToolLink(axes=['x', 'y'], boundsx=(0, np.float64(41.6625)), boundsy=(-0.5, 21), intervalsx=None, intervalsy=None, inverted=True, name='RangeToolLink00869', start_gesture='tap', use_handles=False)
Finally, we’ll arrange the main plot and minimap into a single column layout.
app = (curves_overlay_lttb + minimap).opts(shared_axes=False).cols(1)
It is recommended to only have a single rendered instance of the same application component in a notebook, so we’ve commented out the following cell so we can demonstrate additional extensions below. But feel free to uncomment and display the following if you don’t want any further extensions.
# app
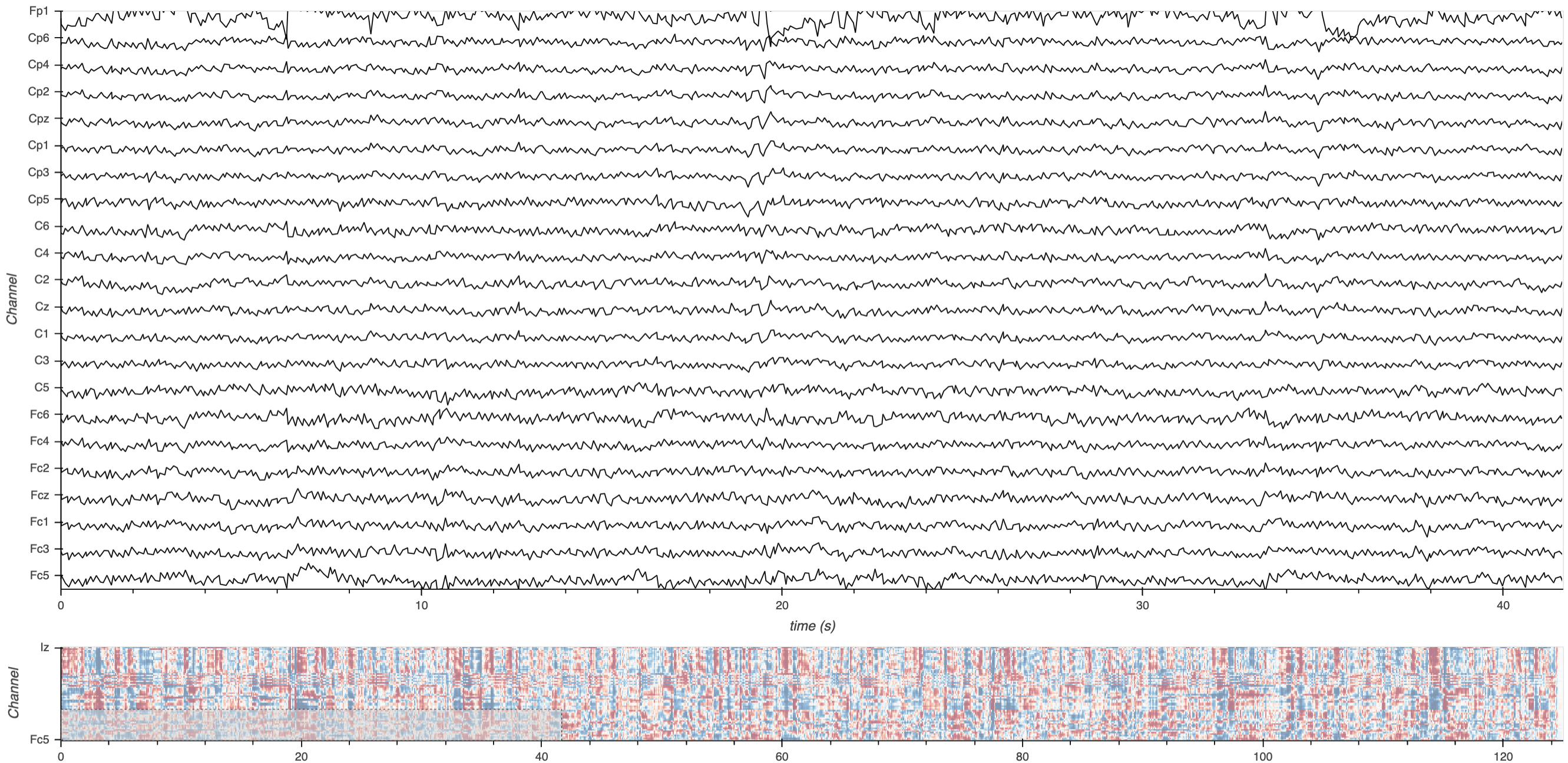
Here’s a static snapshot of what the previous cell produces (a layout of two DynamicMap elements - the downsampled curves plot and a rasterized minimap plot). 👉
# print(app)
Scale Bar Extension#
While the hover tooltips in our plot allow us to inspect amplitude values at specific points, sometimes it’s beneficial to have a persistent visual reference for amplitude measurements. Adding a scale bar provides a constant guide to the amplitude scale, enhancing the interpretability of our visualization, especially when comparing signal amplitudes across different channels or time windows.
In HoloViews, we can incorporate a scale bar into any curve element. This feature displays a bar that represents a specific amplitude value, which can be toggled on or off using the measurement ruler icon in the plot’s toolbar. This allows users to choose whether they want the scale bar visible.
For our application, we’ll add a scale bar to one of the EEG channels—specifically, channel 'C1'. We’ll customize the scale bar’s appearance and behavior using .opts, ensuring it aligns with the units of our data (microvolts, µV). The scalebar_opts dictionary allows us to fine-tune the visual aspects of the scale bar, such as line width, label location, background color, and whether the scale bar size adapts when zooming. See more options in the Bokeh docs.
curves['C1'].opts(
scalebar=True,
scalebar_location="right",
scalebar_unit=('µV', 'V'),
scalebar_opts={
'bar_line_width':3,
'label_location':'left',
'background_fill_color':'white',
'background_fill_alpha':.6,
'length_sizing':'adaptive',
'bar_length_units': 'data',
'bar_length':.8,
}
); # add semicolon to prevent display of output
Uncomment and display the following if you don’t want any further extensions.
# app
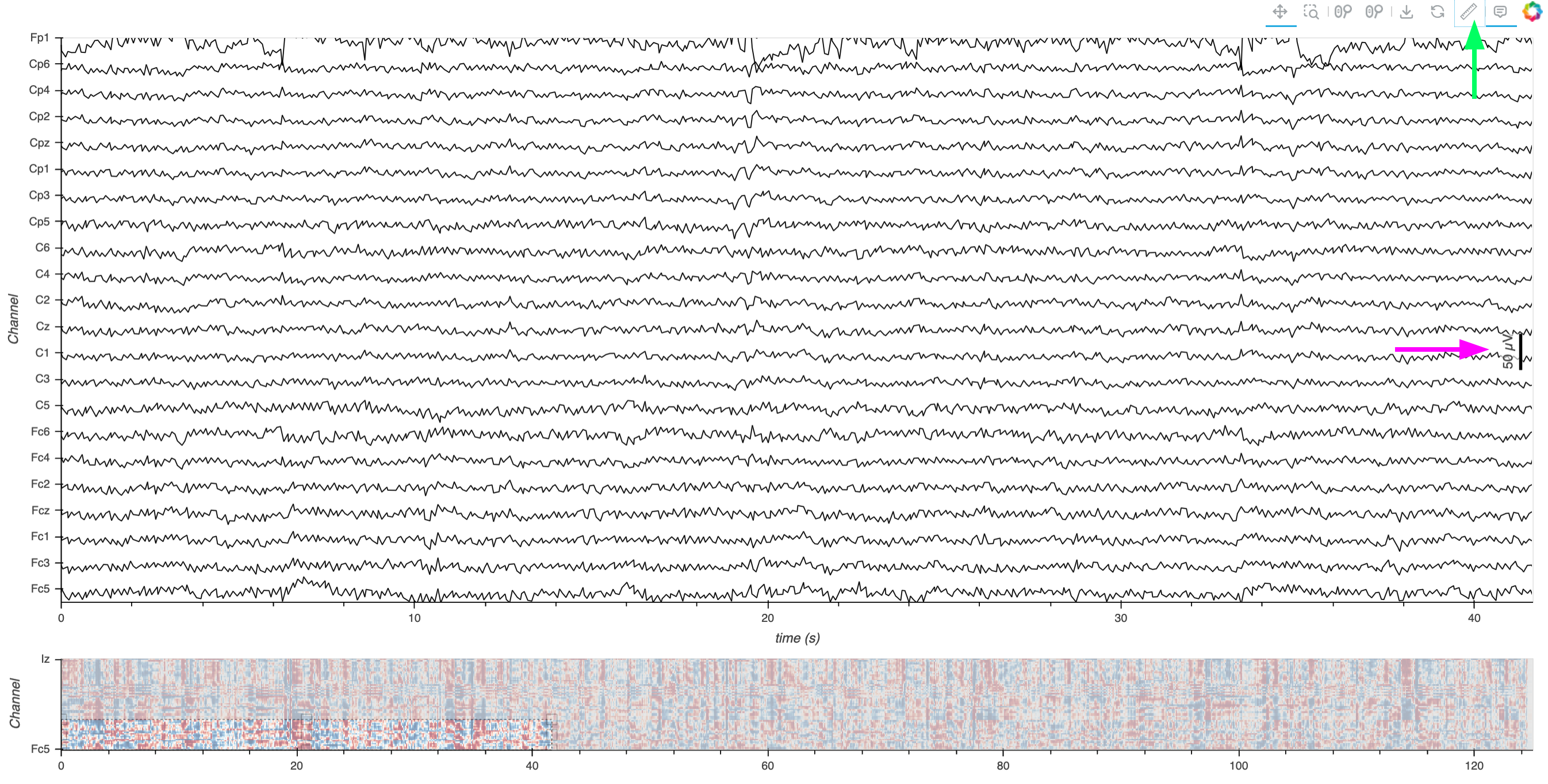
Here’s a static snapshot of what the previous cell produces (the previous layout with the addition of a scalebar for one of the curves). 👉
The pink arrow annotation indicates the newly added adaptive scale bar, and the green arrow points to the toolbar icon used to toggle its display.
# print(app)
As you interact with the plot by zooming in and out, you’ll notice that the scale bar adapts to the current zoom level. This dynamic adjustment ensures that the scale bar remains a relevant reference regardless of the level of detail you’re viewing.
Time Range Annotation Extension#
In this extension, we’ll enhance our multichannel timeseries visualization by adding interactive time range annotations. This feature allows us to mark and label specific periods within the data, which can be invaluable for highlighting events, artifacts, or regions of interest during analysis.
We’ll utilize the HoloNote package, which extends HoloViews with tools for annotating plots and integrating them into interactive dashboards.
Preparing Annotations Data#
If you don’t have any preexisting annotations, that’s perfectly fine. You can proceed without an initial DataFrame and use HoloNote to create annotations from scratch interactively within your visualization. In this case, you can initialize an empty DataFrame or skip the DataFrame preparation step.
However, if you do have preexisting annotations, either from a separate file or embedded in the dataset, read them into a Pandas DataFrame as we do below and adjust the code accordingly to fit your data structure. We’ll demonstrate how to extract annotations that are embedded in our EEG demo dataset (See men.Annotations for information on handling these annotations):
if any(raw.annotations):
# Get the initial time of the experiment
orig_time = raw.annotations.orig_time
# Convert the annotations to a DataFrame
annotations_df = raw.annotations.to_data_frame()
# Ensure the 'onset' column is in UTC timezone
annotations_df['onset'] = annotations_df['onset'].dt.tz_localize('UTC')
# Create 'start' and 'end' columns in seconds relative to the experiment start
annotations_df['start'] = (annotations_df['onset'] - orig_time).dt.total_seconds()
annotations_df['end'] = annotations_df['start'] + annotations_df['duration']
# Preview the annotations DataFrame
annotations_df.head()
else:
# If no annotations are included in our dataset, initialize with an empty data
annotations_df = pd.DataFrame(columns=['start', 'end'])
Setting Up the Annotator#
Now we’ll configure the Annotator object from HoloNote, which manages the annotations and integrates them with our plots.
In this demo, we’re using a connector to an in-memory SQLite database by specifying SQLiteDB(filename=':memory:') for temporary storage of annotations:
from holonote.annotate import Annotator
from holonote.app import PanelWidgets, AnnotatorTable
from holonote.annotate.connector import SQLiteDB
annotator = Annotator(
{'time': float}, # Indicates that we are working with time-based annotations
fields=["description"],
groupby = "description", # Group annotations to enable color-coding and batch actions
connector=SQLiteDB(filename=':memory:'),
)
Note on Persistent Storage: If we don’t specify a connector, HoloNote will by default create a persistent SQLite database file named ‘annotations.db’ in the current directory. You can also specify a custom filename in place of ':memory:' to store annotations persistently in a specific file, such as SQLiteDB(filename='my_annotations.db').
When working in a real workflow, you’ll likely want persistent storage to save your annotations between sessions. Be aware that if a file with the specified name already exists, HoloNote will use it, and any changes committed through the UI will modify this file. This allows you to maintain and update your annotations across different sessions, ensuring your work is saved and accessible later.
Defining Annotations#
We now populate the Annotator with the annotations from our DataFrame. If you have an existing annotations_df, whether extracted from the dataset or loaded from a file, you can define the annotations as follows:
annotator.define_annotations(annotations_df, time=("start", "end"))
This line tells the annotator to use the ‘start’ and ‘end’ columns from our DataFrame as the time ranges for the annotations.
If you started without any existing annotations, you can skip this step, and the Annotator will start without any annotations, ready for you to add new ones interactively.
Configuring Annotation Widgets#
To enable interaction with the annotations, we’ll create widgets that allow users to view and manage them.
annotator.visible = ["T1", "T2"] # Optional, specify which to initially display
anno_controls = PanelWidgets(annotator)
anno_table = AnnotatorTable(
annotator,
tabulator_kwargs=dict(
pagination="local",
page_size=10,
sizing_mode="stretch_both",
layout="fit_columns",
theme="default",
stylesheets=[":host .tabulator {font-size: 12px;}"],
text_align={"description": "center"},
),
)
annotator_widgets = pn.WidgetBox(
anno_controls,
anno_table,
max_width=325,
)
The above code creates a WidgetBox containing:
Annotation controls (PanelWidgets) for adding, editing, or deleting annotations. Since we grouped the annotations,
PanelWidgetswill also include a ‘Visible’ widget to toggle visibility by group.An
AnnotatorTableto display annotations in a tabular format.
We also customize the table’s appearance and functionality using tabulator_kwargs, adjusting pagination, sizing, layout, theme, style, and text alignment. Check our the Panel Tabulator docs for more options.
Integrating Annotations with the Plots#
Finally, we’ll overlay the annotations onto our existing multichannel timeseries plot and the minimap. This integration allows us to see the annotations directly on the plots and interact with them.
Add new annotations by:
Ensure that the ‘+’ button is selected in the widgets.
Ensure that ‘Box select (x-axis)’ is selected in the Bokeh Toolbar.
Enter the name of the group that you want the annotation to be added to in the ‘description’ text box.
Select a time range by clicking and dragging on the plot
Edit or delete annotations using the provided widgets and table.
Note, if you are using a persistent file on disk for the annotations, hit the triangle button to commit changes to the file.
annotated_multichan = (annotator * curves_overlay_lttb).opts(responsive=True, min_height=700)
annotated_minimap = (annotator * minimap)
annotated_app = (annotated_multichan + annotated_minimap).cols(1).opts(shared_axes=False)
⚠️ Heads up!
Viewing this on examples.holoviz.org? This is a static version of the notebook 📄. Interactive features that require a live Python process, like downsampling and datashading, won’t work here 🚫. To use these features, open the notebook with a live Python session.
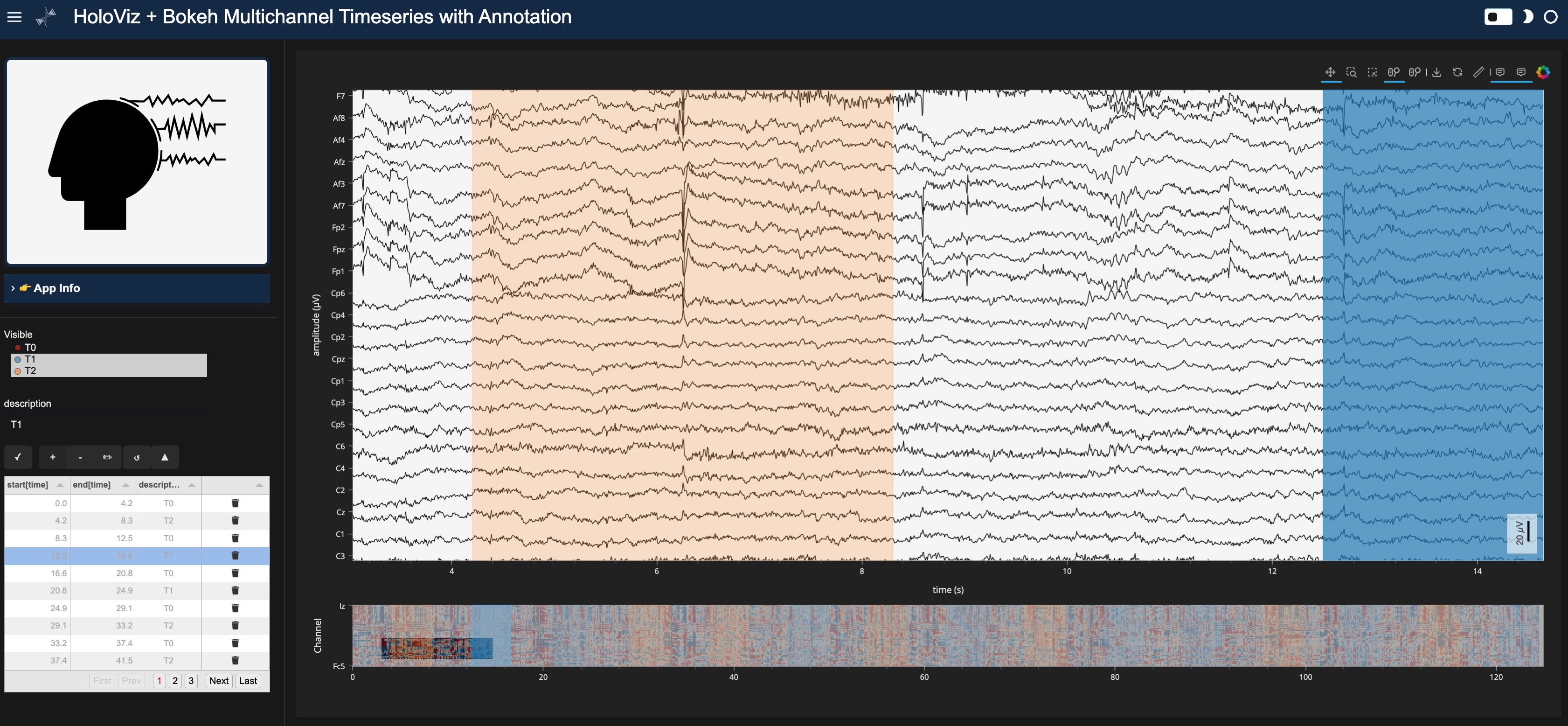
pn.Row(annotator_widgets, annotated_app)
Standalone App Extension#
HoloViz Panel allows for the deployment of this complex visualization as a standalone, template-styled, interactive web application (outside of a Jupyter Notebook). Read more about Panel here.
We’ll add our plots to the main area of a Panel Template component and the widgets to the sidebar. Finally, we’ll set the entire component to be servable.
To launch the standalone app, activate the same conda environment and run panel serve <path-to-this-file> --show in the command line. This will open the application in a browser window.
Warning
It is not recommended to have both a notebook version of the application and a served of the same application running simultaneously. Prior to serving the standalone application, clear the notebook output, restart the notebook kernel, and saved the unexecuted notebook file.app_info = pn.Card(
pn.pane.Markdown(
"""
**For guidance, visit the [Workflow](https://examples.holoviz.org/gallery/multichannel_timeseries/0_multichan.html).**
This app demonstrates interactive viz of [multichannel timeseries data](https://examples.holoviz.org/gallery/multichannel_timeseries/index.html) using [HoloViz](https://holoviz.org) and [Bokeh](https://bokeh.org/).
Ideal for analyzing and annotating electrophysiological data like EEG, ECoG, or LFP recordings that fit in memory.
For handling larger datasets that exceed memory, see this [other Workflow](https://examples.holoviz.org/gallery/multichannel_timeseries/1_large_multichan.html).
""",
sizing_mode="stretch_width",
styles={"color": "black",
},
),
title="👉 App Info",
sizing_mode="stretch_width",
header_background="#122944",
header_color="WhiteSmoke",
styles={"background": "WhiteSmoke",
},
margin=10,
collapsed=True,
)
eeg_svg = pn.Column(
pn.pane.SVG(
"https://raw.githubusercontent.com/holoviz-topics/examples/e9e3346e619671d23e2a0511ae762b8b33ff0ddc/multichannel_timeseries/assets/eeg.svg",
sizing_mode="scale_both",
),
sizing_mode='stretch_both',
min_height=300,
styles={"background": "WhiteSmoke",
"border-radius": "10px",
"border": "4px solid #122944"},
margin=10,
)
servable_app = pn.template.FastListTemplate(
site_url="https://holoviz.org",
logo="https://raw.githubusercontent.com/holoviz/holoviz/refs/heads/main/doc/_static/holoviz-logo.svg",
favicon="https://raw.githubusercontent.com/holoviz/holoviz/refs/heads/main/doc/_static/favicon.ico",
sidebar=[eeg_svg, app_info, pn.layout.Divider(), anno_controls, anno_table],
title="HoloViz + Bokeh Multichannel Timeseries with Annotation",
main=annotated_app,
accent='#122944',
theme='dark',
sidebar_width=400,
).servable()
Here’s a static snapshot of what the previous cell produces in a browser window when you serve the standalone app (a templated Panel application). 👉
print(servable_app)
<FastListTemplate FastListTemplate05092>
What Next?#
Head back to the Index page to explore other workflow alternatives, such as Large Multichannel Timeseries.